International Journal of Veterinary Science and Research
Structural characteristics of the control region of the Beaufortia kweichowensis mitochondrial genome
Gang Wang1, Gui-Hong Chen1, Yu Luo2 and Zheng-Yong Wen1*
2Institute of Aquaculture, Neijiang Academy of Agricultural Sciences, Neijiang, Sichuan 641000, China
Cite this as
Wang G, Chen GH, Luo Y, Wen ZY (2018) Structural characteristics of the control region of the Beaufortia kweichowensis mitochondrial genome. Int J Vet Sci Res 4(1): 012-014. DOI: 10.17352/ijvsr.000029In present study, we identified the structural characteristics of the Beaufortia kweichowensis mtDNA control region using the next-generation sequencing method. Result showed that the control region could be further divided into three parts, including the extended termination associated sequence domain (ETAS), the central conserved domains (CSB-F, CSB-E, CSB-D) and the conserved sequence block domains (CSB-1, CSB-2, CSB-3), and their conserved sequences were identified. Additionally, we found the ETAS domain could be folded and form a 25 bp loop, which are usually considered to play an important role in mtDNA transcript termination. Finally, we reconstructed the phylogenetic relationship among the family of Balitoridae based on the nucleotide sequence of the control region, result showed that the Balitoridae can be divided into two clades, one clade is Gastromyzoninae and the other is Homalopterinae, and the B. kweichowensis was grouped into clade Gastromyzoninae and shared a close relationship with B. szechuanensis.
Introduction
Mitochondrial DNA (mtDNA) is a circular double-strand DNA with a 15-20 kb length in animals, and it usually encodes 37 genes, including 13 protein-coding genes (PCGs), 22 transport RNA (tRNA), and 2 ribosomal RNAs (rRNAs) [1,2]. To date, mtDNA has been widely used for genetic research, taxon classification, phylogenetic evolution research and population studies attribute to its fast variation, maternal inheritance, rapid evolution and lack of recombination [3,4]. Recent years, the mitochondrial genome database growth rapidly following the fast development of high throughput sequencing technology [5], which might provide more chance for scientists to solve the biological mystery in the future.
The control region, also called displacement-loop region (D-loop), is a DNA fragment with fastest evolution rate in mtDNA because it composes of the single strand nucleotide [6]. It can be divided into three domains, including the extended termination associated sequence domain (ETAS), the central conserved domains (CSB-F, CSB-E, CSB-D) and the conserved sequence block domains (CSB-1, CSB-2, CSB-3) [7,8], and it also has been extensively used for phylogenetic analyses and populational genetics researches [9,10].
Fish is one of the most amount groups in vertebrate, and now more than 30000 species has been discovered [11]. Thus far, more than 200 fish genome and 1000 fish mitochondrial genome has been sequenced and deposited into NCBI database, and these data are available for all researchers, thus then some important scientific issue such as the evolutionary relationship of fish has been well investigated [12,13], which also will be helpful for species classification and phylogenetic analyses in these groups.
B. kweichowensis (Fang) is an endemic fish of the upper reaches of the Beipan River of China, belonging to the family of Balitoridae (Cypriniformes), has not yet been well studied about its phylogenetic evolutionary status [2]. In present study, the structure of control region of B. kweichowensis mtDNA was investigated, and the conserved sequences also was identified. Subsequently, the phylogenetic tree of the family Balitoridae was reconstructed based on the sequence information of mtDNA control region, which could be helpful to establish the evolutionary status of this species.
Material and Methods
Sample collection and DNA isolation
One adult B. kweichowensis used in this study were obtained from Conservation and Utilization of Fishes resources in the Upper Reaches of the Yangtze River Key Laboratory of Sichuan Province, Neijiang Normal University. A 20–30 mg fin clip was collected and preserved in 95% ethanol at 4 °C. Total genomic DNA was extracted with a Tissue DNA Kit (OMEGA E.Z.N.A.) following the manufacturer’s protocol.
Sequencing and structure analysis of control region
The genomic DNA was sequenced using the next-generation sequencing, and then the mitogenome was assembled using B. szechuanensis (GenBank accession number: KP716708.1) as reference. The non-coding control region structure was analyzed and drawn according to the control region structure of other Botiinae fish.
Phylogenetic analysis
The phylogenetic tree was constructed with nucleotide sequence of control region using MEGA 6.0 software [14]. The HKY+G model was selected as the best evolutionary model after testing, and the tree was performed using maximum likelihood and bootstrapped with 1000 replications. The species Cycleptus elongatus and Catostomus commersonii were selected as outgroups.
Results and Discussion
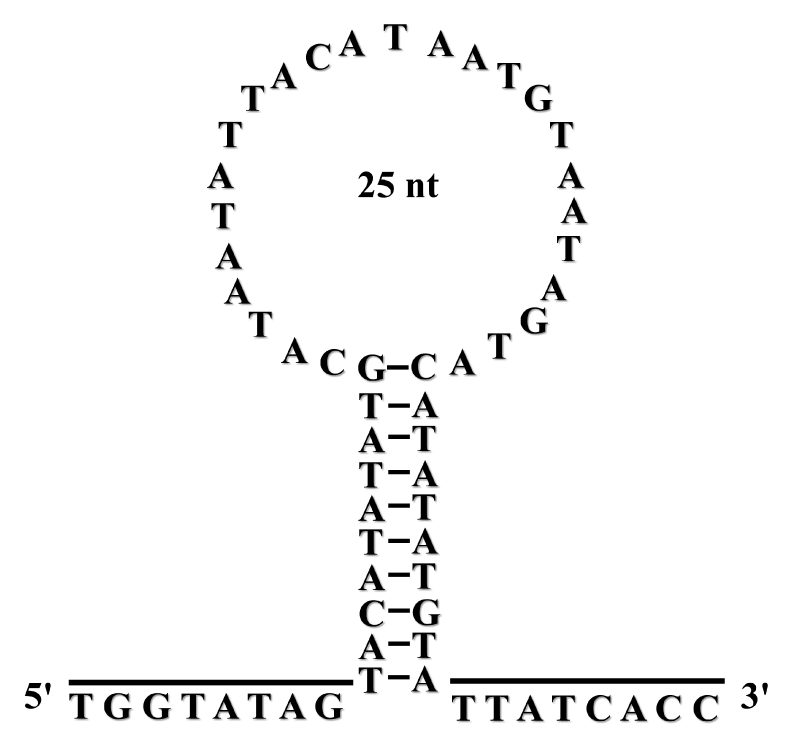
The full length of the control region of B. szechuanensis mtDNA is 930 bp in length, and it can be divided into three domains including the extended termination associated sequence domain (ETAS), the central conserved domains (CSB-F, CSB-E, CSB-D) and the conserved sequence block domains (CSB-1, CSB-2, CSB-3), result is shown in figure 1. Simultaneously, the conserved sequences of these domains were identified, the conserved sequence of ETAS is CATATATGCATAATATTACATAATGTAATAGTACATATATGTA, and it can be folded into a 25 bp long loop structure (Figure 2), which is usually considered to be connection with mitochondria transcript termination [2], and this phenomenon also can be found in other fishes [15]. The conserved sequences of the central conserved domains are ATGTAGTAAGAAACCACCAACCAGTTTA, TCAGGGACAATAATCGTGGGGG and TGAATTATTACTGGCATCTGGTTCCTATTTCAGG, which represent the conserved sequence of CSB-F, CSB-E, CSB-D domains, respectively. Moreover, the conserved sequence of the conserved sequence block domains is AGGTTAATGATTAAATGACATAACTCAAGA, CGCGAGAGGCCCCCTTACCCCCTTACAC and CCTTGTCAAACCCCGAAACCAAGGAA, which represent the conserved sequence of CSB-1, CSB-2, CSB-3 domains, respectively. Furthermore, the TATA element also was identified, which is considered to play an important role in transcript origination [9].
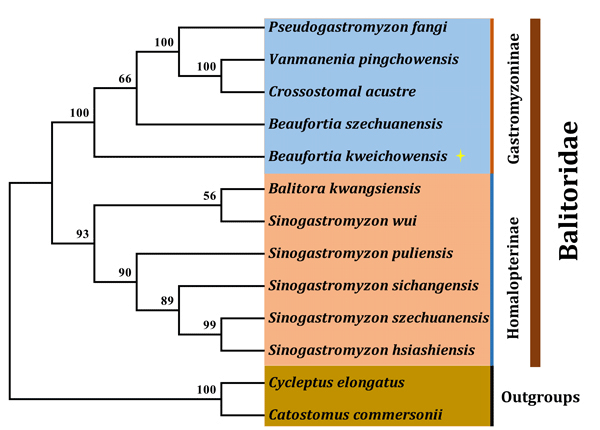
The Balitoridae is regarded as a distinct family in order Cypriniformes. Hora divided this family into two subfamilies Gastromyzoninae and Homalopterinae, and he considered the former as derivative of the Cobitidae and the latter as a descendant of the family Cyprinidae [16]. In order to better understand the relationship of this family, maximum likelihood phylogenetic tree was reconstructed based on the nucleotide sequence of mtDNA. Results showed that the family Balitoridae exactly can be divided into two clades, including subfamily Gastromyzoninae and Homalopterinae, and the B. kweichowensis was grouped into clade Gastromyzoninae and shared a close relationship with B. szechuanensis (Figure 3). These findings are consistent with our previous study based on whole mtDNA sequence, suggesting the B. kweichowensis should be origin from the subfamily Gastromyzoninae [17].
Conclusion
The control region of B. kweichowensis could be divided in two three domains, and their conserved sequence were identified. Furthermore, a 25 bp long loop domain was found in the ETAS. Finally, the phylogenetic relationship of the family Balitoridae was analyzed, result highly supported the B. kweichowensis belongs to the subfamily Gastromyzoninae.
This work was supported by the Scientific Research Fund of Sichuan Education Department (No. 18ZB0328; No. KYTD201009) and the patent research program of Neijiang Normal University (No. 17ZL03).
- Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767-1780. Link: https://tinyurl.com/yd2rn675
- Zou YC, Xie BW, Qin CJ, Wang YM, Yuan DY, et al. (2017) The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genomics 39: 767-778. Link: https://tinyurl.com/yafh9n8y
- Zou YC, Chen M, Qin CJ, Wang YM, Li R, et al. (2017) Identification of the complete mitochondrial genome of Garra pingi pingi (Cypriniformes, Cyprinidae). Conserv Genet Resour 1-4. Link: https://tinyurl.com/y7qx3g7o
- Miller W, Drautz DI, Janecka JE, Lesk AM, Ratan A, et al. (2009) The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Res 19: 213-220. Link: https://tinyurl.com/y98c233e
- Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11: 31-46. Link: https://tinyurl.com/y76rfgqz
- Zhao JL, Wang WW, Li SF, Cai WQ (2006) Structure of the mitochondrial DNA control region of the sinipercine fishes and their phylogenetic relationship. Acta Genet Sin 33: 793-799. Link: https://tinyurl.com/yd54xqhz
- Southern SO, Southern PJ, Dizon AE (1988) Molecular characterization of a cloned dolphin mitochondrial genome. J Mol Evol 28: 32-40. Link: https://tinyurl.com/yabnf9ey
- Sbisa E, Tanzariello F, Reyes F, Pesole G, Saccone C (1997) Mammalian mitochondrial D-loop region structure analysis: identification of new conserved sequences and the functional and evolutional implications. Gene 205: 125-140. Link: https://tinyurl.com/ydec29yx
- Lee WJ, Conroy J, Howell WH, Kocher TD (1995) Structure and evolution of teleost mitochondrial control regions. J Mol Evol 41: 54-56. Link: https://tinyurl.com/ya9omwg2
- Guo XH, Liu SJ, Liu Y (2003) Comparative analysis of the mitochondrial DNA control region in cyprinids with different ploidy level. Aquaculture 224: 25-38. Link: https://tinyurl.com/y8fbockp
- Casal CMV (2006) Global documentation of fish introductions: the growing crisis and recommendations for action. Bioinvasions Rec 8: 3-11. Link: https://tinyurl.com/y7qy6vrh
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, et al. (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A 109: 13698-13703. Link: https://tinyurl.com/y6ubmo3j
- Hughes LC, Ortí G, Huang Y, Sun Y, Baldwin CC, et al. (2018) Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci U S A115: 6249-6254. Link: https://tinyurl.com/yb96tb89
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725-2729. Link: https://tinyurl.com/yandh6pl
- Mu X, Liu Y, Lai M, Song H, Wang X, et al. (2015) Characterization of the Macropodus opercularis complete mitochondrial genome and family Channidae taxonomy using Illumina based de novo transcriptome sequencing. Gene 559: 189-195. Link: https://tinyurl.com/y8x3hbnl
- Hora S (1932) Classification, bionomics and evolution of Homalopteridae fishes. Mem Indian Mus 12: 263-330. Link: https://tinyurl.com/ya4uss78
- Wen ZY, Xie BW, Qin CJ, Wang J, Yuan DY, et al. (2017) The complete mitochondrial genome of a threatened loach (Beaufortia kweichowensis) and its phylogeny. Conserv Genet Resour 9: 565-568. Link: https://tinyurl.com/y8xgbefj
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
